Cancer
The Berger Lab cancer group currently has two main areas of interest: the molecular mechanisms of the p53 protein family, and the epigenetics of T cell dysfunction.
Epigenetic mechanisms of the p53 family
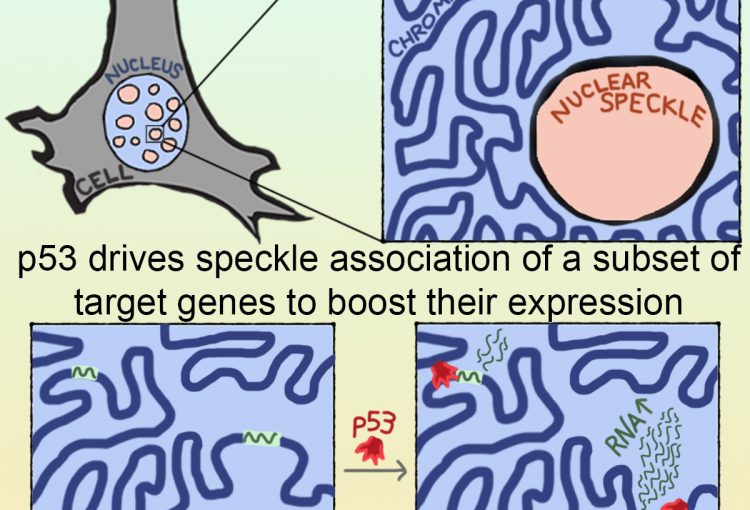
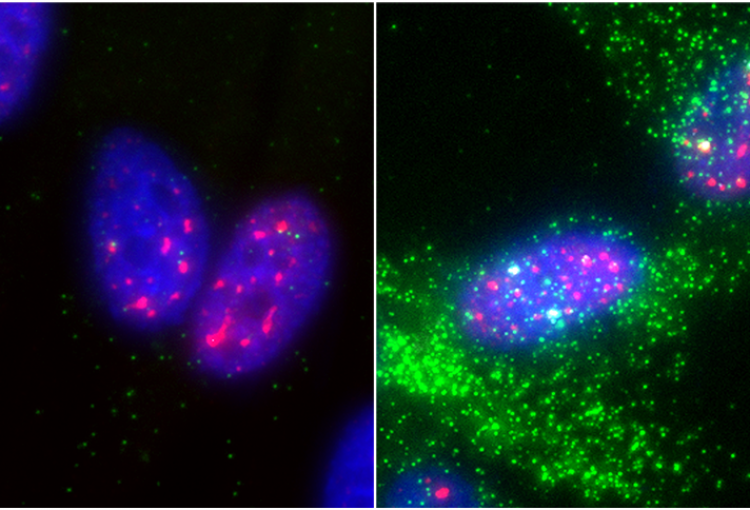
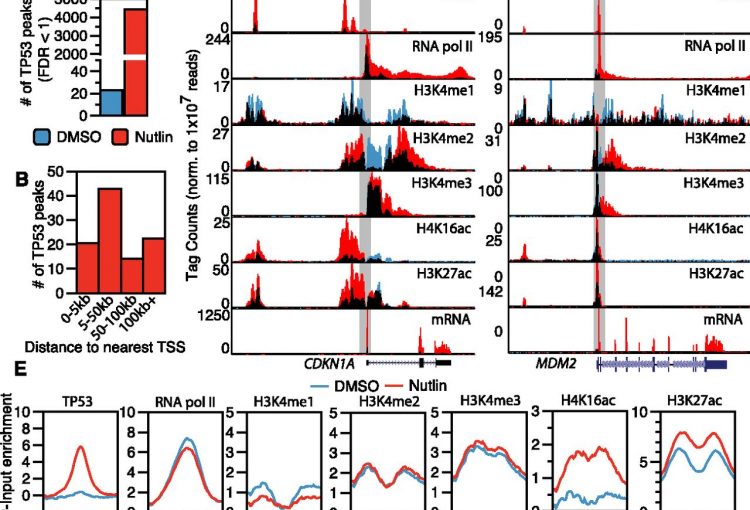
The molecular mechanisms of the p53 protein family are a long-standing interest of the Berger Lab. Being a critical tumor suppressor protein, wild type p53 is a key barrier to oncogenic transformation. On the other hand, p53 often acquires point mutations that shift p53’s tumor suppressive functions towards oncogenic functions, harnessing p53’s potent transactivation abilities to induce expression of genes that promote cancer rather than prevent it. In the lab, we investigate the chromatin mechanisms of wild type and mutant p53 and how they impart tumor suppressive or oncogenic functions. These studies encompass the genomic binding strategies of wild type and mutant p53, their relationship with histone modifications, post-translational modification of p53, and, more recently, the ability of p53 to modulate nuclear localization of its target genes in relation to activating nuclear bodies.
The p63 protein is a close family member of p53. However, in contrast to p53, p63 is essential in mammals and is a key mediator of epidermal development. Much as with our studies of p53, we seek to link the chromatin functions of p63 to disease outcomes. In particular, through our combination of epigenomic and GWAS studies, we have identified a strong molecular link between p63 enhancer functions and clefting of the lip/palate, illuminating molecular mechanisms underlying this developmental defect and revealing vital regulatory elements and new candidate causative genes.
Immunoepigenetics
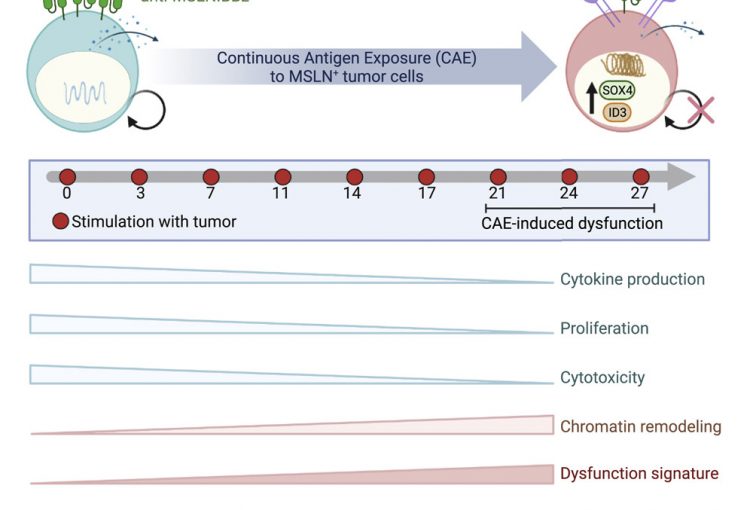
During the course of a chronic infection or cancer, T cells can lose their ability to clear antigen or tumor cells and become exhausted over time. Current cancer therapeutics, such as chimeric antigen receptor (CAR) T therapy and immune checkpoint (PD-1/PD-L1) blockade, are focused on reinvigorating T cell function. In collaboration with Carl June and E. John Wherry, we are trying to dissect the epigenetic state of CAR T cells in cancer patients and during T cell exhaustion in chronic infection, using state of the art epigenetic and genomic techniques. The ultimate goal is to manipulate certain epigenetic pathways in combination with immune checkpoint blockade to reboot exhausted T cell function.
CAR-T cell research
CAR T cell therapy has achieved remarkable success in hematological malignancies, with a 90% complete response rate observed in leukemia. However, this therapy has not worked well in solid tumors. We, in collaboration with Dr. Carl June’s group, have devised an in vitro model to study mechanisms mediating CAR T cell dysfunction against solid tumors, including pancreatic cancer. We use genomic, epigenomic and single cell technologies to investigate the gene expression and epigenetic changes that occur as CAR T cells lose effector function. The ultimate goal is to identify transcription factors and/or small molecule inhibitors that regulate the CAR T dysfunction program. These factors can then be engineered using CRISPR Cas9 technology to be expressed or knocked out in CAR T cells, which holds promise for the development of even more effective CAR T cell therapies designed to treat solid tumors.
T cell exhaustion
CD8+ T cells play a key role in the immune response against intracellular pathogens and tumors. During chronic infections or cancer, CD8+ T cells are continuously exposed to antigen and to inflammatory signals. This constant stimulation leads CD8+ T cells to differentiate into an exhausted state, characterized by loss of cytokine production (IL-2, IFNg, GzmB, TNFa), loss of killing function, expression of inhibitory receptors (PD-1, CTLA4, Tim3, etc.), poor memory potential, and compromised proliferation. In collaboration with Dr. John Wherry, our lab has shown that exhausted CD8+ T cells (Tex) are also characterized by a unique transcriptional program and epigenetic landscape that distinguishes them from normal effector (Teff) or memory (Tmem) CD8+ T cells. Using a combination of genomic approaches and domain targeted CRISPR screenings, we investigate the epigenetic mechanisms leading to exhaustion and how we can modulate these epigenetic pathways to improve immunotherapy and efficiently reinvigorate exhausted cells.
Publications
Genetics Meets Epigenetics in Treg Cells and Autoimmunity. Agudelo Garcia PA, Berger SL. Immunity. 2020 Jun 16;52(6):897-899. doi: 10.1016/j.immuni.2020.05.009. PMID: 32553177
p53 mediates target gene association with nuclear speckles for amplified RNA expression. Alexander, K.A., Cote, A., Nguyen, S. C., Zhang, L., Gholamalamdari, O., Agudelo-Garcia, P., Lin-Shiao, E., Tanim KM. A., Lim, J., Biddle, N., Dunagin, M. C., Good, C.R., Mendoza, M.R., Little, S.C., Belmont, A., Joyce, E.F., Raj, A., Berger, S.L. (2021). Molecular Cell, 81(8):1666-1681.e6.
p63 establishes epithelial enhancers at critical craniofacial development genes. Lin-Shiao, E., Lan, Y., Welzenbach, J., Alexander, K. A., Zhang, Z., Knapp, M., Mangold, E., Sammons, M., Ludwig, K. U., & Berger, S. L. (2019). Science advances, 5(5), eaaw0946.
TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Khan, O., Giles, J. R., McDonald, S., Manne, S., Ngiow, S. F., Patel, K. P., Werner, M. T., Huang, A. C., Alexander, K. A., Wu, J. E., Attanasio, J., Yan, P., George, S. M., Bengsch, B., Staupe, R. P., Donahue, G., Xu, W., Amaravadi, R. K., Xu, X., Karakousis, G. C., Tara C., Mitchell, Lynn M. Schuchter, Jonathan Kaye, Shelley L. Berger, Wherry, E. J. (2019). Nature, 571(7764), 211–218. https://doi.org/10.1038/s41586-019-1325-x
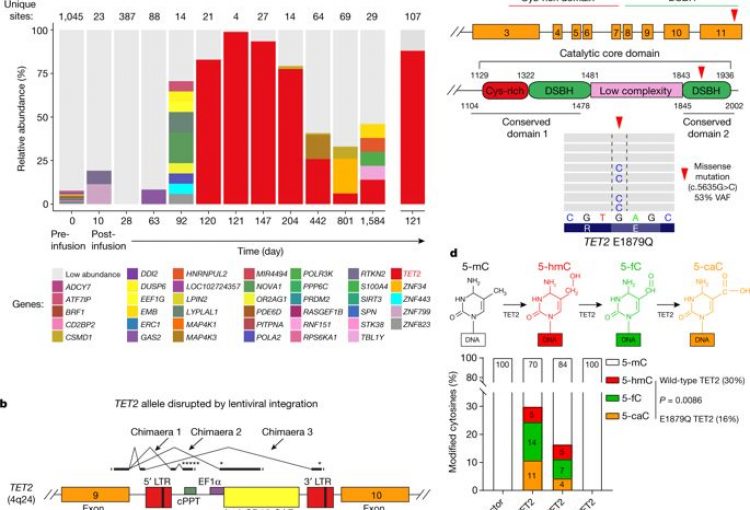
Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Fraietta, J. A., Nobles, C. L., Sammons, M. A., Lundh, S., Carty, S. A., Reich, T. J., Cogdill, A. P., Morrissette, J., DeNizio, J. E., Reddy, S., Hwang, Y., Gohil, M., Kulikovskaya, I., Nazimuddin, F., Gupta, M., Chen, F., Everett, J. K., Alexander, K. A., Lin-Shiao, E., Gee, M. H., Xiaojun Liu, Regina M. Young, David Ambrose, Yan Wang, Jun Xu, Martha S. Jordan, Katherine T. Marcucci, Brice L. Levine, K. Christopher Garcia, Yangbing Zhao, Michael Kalos, David L. Porter, Rahul M. Kohli, Simon F. Lacey, Shelley L. Berger, Frederic D. Bushman, Carl H. June, Melenhorst, J. J. (2018). Nature, 558(7709), 307–312.
Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Pauken, K. E., Sammons, M. A., Odorizzi, P. M., Manne, S., Godec, J., Khan, O., Drake, A. M., Chen, Z., Sen, D. R., Kurachi, M., Barnitz, R. A., Bartman, C., Bengsch, B., Huang, A. C., Schenkel, J. M., Vahedi, G., Haining, W. N., Berger, S. L., & Wherry, E. J. (2016). Science (New York, N.Y.), 354(6316), 1160–1165.
Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Zhu, J., Sammons, M. A., Donahue, G., Dou, Z., Vedadi, M., Getlik, M., Barsyte-Lovejoy, D., Al-awar, R., Katona, B. W., Shilatifard, A., Huang, J., Hua, X., Arrowsmith, C. H., & Berger, S. L. (2015). Nature, 525(7568), 206–211.
Lysine methylation represses p53 activity in teratocarcinoma cancer cells. Zhu, J., Dou, Z., Sammons, M. A., Levine, A. J., & Berger, S. L. (2016). Proceedings of the National Academy of Sciences of the United States of America, 113(35), 9822–9827. https://doi.org/10.1073/pnas.1610387113