Neuroepigenetics
Adaptation to the environment is a central problem for all organisms, and adaptive behaviors are therefore essential for organismal life. Epigenetic mechanisms, notably DNA methylation, post-translational histone modification, and chromatin remodeling have been shown to regulate environmentally-responsive dynamic gene expression in the nervous systems of many organisms, facilitating adaptive behavioral repertoires. However, specific pathways responsible for the establishment, maintenance, and modification of neuronal gene expression patterns related to behavioral adaptation remain elusive. To address this problem, our lab studies behavioral epigenetics using two disparate and complementary systems: mammals and ants.
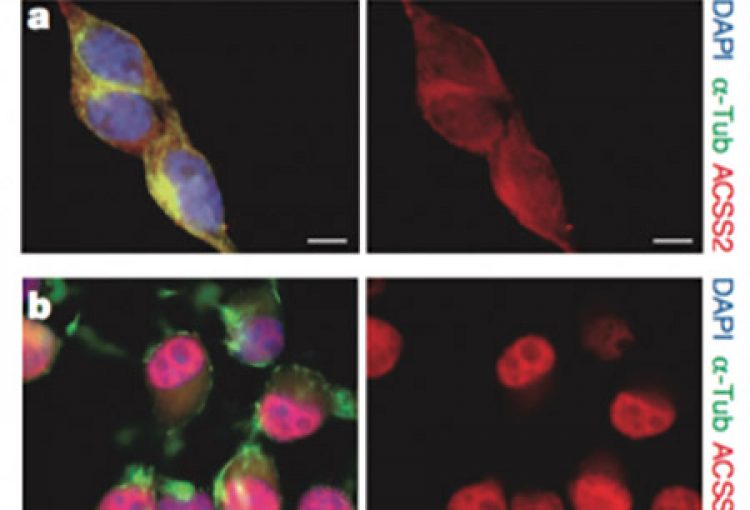
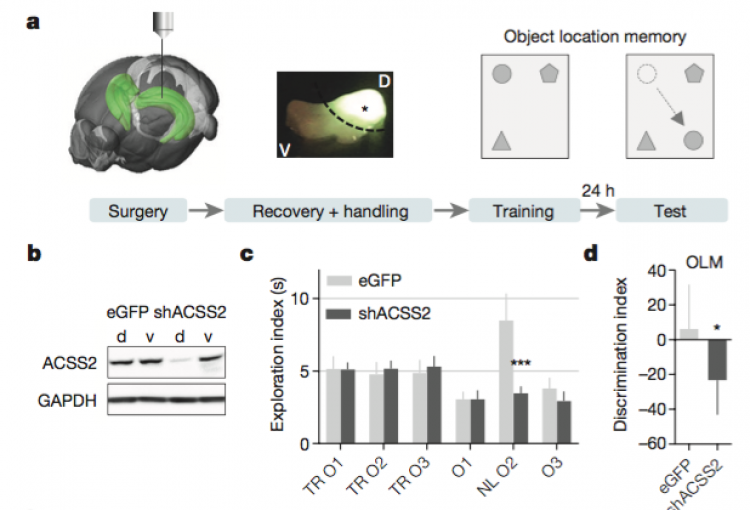
In mammals, memory formation is a central component of virtually all complex behaviors. This process involves plasticity of neuronal synapses, which is in part driven by activity-dependent transcriptional programs regulated by epigenetic mechanisms. Our lab has recently identified a strong connection between neuronal plasticity, epigenetics and cellular metabolism. In particular, we have shown that ACSS2 (acelyl-CoA synthetase 2) acts as a chromatin-bound transcriptional co-activator in hippocampal neurons, a brain area required for the formation of spatial memories. During formation and reconsolidation of spatial object memory, ACSS2 drives histone acetylation and transcription of immediate early genes with key functions in neuronal plasticity. This is achieved by the tight regulation of local acetyl-CoA levels that in turn fuel histone acetyltransferase activity. Strikingly, while dorsal hippocampal ACSS2 knock-down did not cause gross behavioral alterations, it specifically ablated spatial object recognition. Our data provide a better understanding for the role of the metabolic-epigenetic axis in memory and have far-reaching consequences for the treatment of cognitive and neuropsychiatric illnesses.
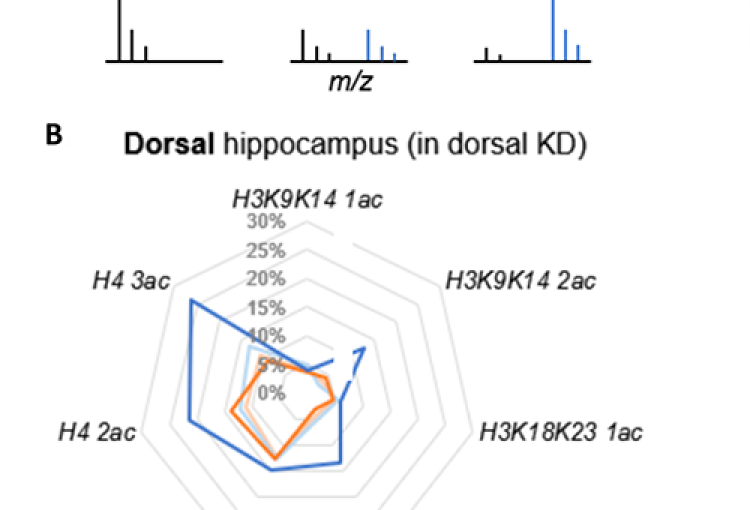
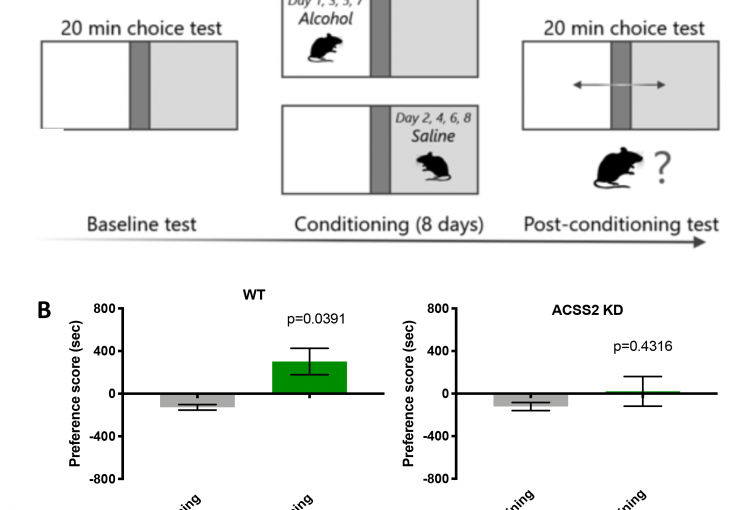
As a major biological source of acetate is alcohol metabolism in the liver, we next explored whether alcohol-derived acetate affects the epigenetic landscape in the brain following binge drinking. Indeed, using heavy isotope labeling in mice, we found that liver alcohol metabolism rapidly fuels histone acetylation in the brain by direct deposition of alcohol-derived acetyl groups onto histones in an ACSS2-dependent manner. In isolated primary hippocampal neurons in vitro and in mouse brain in vivo, acetate induced learning and memory-related transcriptional programs that were sensitive to ACSS2 inhibition. Strikingly, we found that ACSS2 is required for ethanol-induced associative learning, which underlies craving and relapse after protracted periods of abstinence. These findings establish a direct and dynamic link between peripheral alcohol metabolism and neuronal histone acetylation with significant therapeutic potential.
Publications
Food for thought – the nuclear metabolic-epigenetic axis bridges the environment and genes to modulate behavior.
Egervari G, Glastad KM, Berger SL. Science, 2020 Nov;370(6517):660-662
An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease
Nat Genet. 2020 Oct;52(10):1024-1035. doi: 10.1038/s41588-020-0696-0. Epub 2020 Sep 28. PMID: 32989324
Alcohol metabolism contributes to brain histone acetylation
Mews, P., Egervari, G., Nativio, R. et al. Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721 (2019) doi:10.1038/s41586-019-1700-7
Regulation of Chromatin and Gene Expression by Metabolic Enzymes and Metabolites
Li X, Egervari G, Wang Y, Berger SL, Lu Zhimin. Nature Reviews Molecular Cell Biology. 2018 June 21. doi: 10.1038/s41580-018-0029-7.
Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease.
Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F, Toledo J, Gosai S, Gregory BD, Torres C, Trojanowski JQ, Wang LS, *Johnson FB, *Bonini NM, *Berger SL. (2018). Nature Neuroscience, in press. *co-corr authors
Acetyl-CoA Synthetase Regulates Histone Acetylation and Hippocampal Memory
Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL. Nature. 2017 May 31. doi: 10.1038/nature22405.