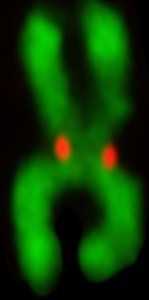
Centromeres
 The realization that the chromosomal locus that drives inheritance of our DNA is defined epigenetically, rather than by a particular DNA sequence, emphasizes how much is left to uncover before we understand the molecular processes guarding our genes each time the cell divides. Building on our many contributions to this area of chromosome biology and the diverse approaches that we have developed and used in recent years, we are attacking the topic of centromeres from many angles. Our overarching strategy is to have our discoveries in one of these areas influence further experimentation in one or more of the other areas.
The realization that the chromosomal locus that drives inheritance of our DNA is defined epigenetically, rather than by a particular DNA sequence, emphasizes how much is left to uncover before we understand the molecular processes guarding our genes each time the cell divides. Building on our many contributions to this area of chromosome biology and the diverse approaches that we have developed and used in recent years, we are attacking the topic of centromeres from many angles. Our overarching strategy is to have our discoveries in one of these areas influence further experimentation in one or more of the other areas.
- Structural biochemistry of the centromere. A deviant nucleosome, containing the histone H3 variant, CENP-A, is the key repeating unit of the chromatin that epigenetically marks centromere location. Using this nucleosome as a point of orientation, we are reconstituting complexes that form at chromatin containing this nucleosome, as well as those involved in the cell cycle-coupled program of epigenetic centromere inheritance for structural and functional studies. For the structural studies, we apply both conventional approaches as well as emerging solution-based methods. One approach that has been particularly helpful at the centromere, is our work with hydrogen/deuterium exchange coupled to mass spectrometry (HXMS). For the functional studies, we use a battery of cell biological approaches at the leading edge of our field to test the hypotheses arising from our structural work.
- Genomics and epigenomics of centromeres. The relationship between the genetic and epigenetic components to centromere identity remains one of the great mysteries in chromosome biology. On one hand, there is a growing list of examples in nature of centromeres that do not require the highly repetitive DNA typically found at this part of the chromosome. On the other hand, human centromeres contain repeats that are highly similar (60-100% sequence identity) on each of our chromosomes, and there are recent observations that these may have a positive impact on centromere function. We are currently using next generation sequencing-based methodologies to interrogate the architecture of centromeric chromatin and examine the DNA sequences wrapping CENP-A nucleosomes.
- Transmitting centromere identity through the mammalian germline. The molecular basis for the transmission of genetic information from each parent has long been a topic of biology textbooks. The molecular basis for transgenerational transmission of epigenetic information largely remains to be defined, despite many documented examples where it surely occurs, including the transmission of centromere location on the chromosome. Joining forces with our close collaborator, Dr. Michael Lampson, and his colleagues in the Dept of Biology here at UPenn, we are using advanced gene targeting approaches to manipulate the epigenetic components in centromere specification. These manipulations are part of tests designed to ask questions in both the female and male mammalian germline. In addition to providing information that will be relevant to understanding multiple forms of transgenerational epigenetic inheritance, these studies are central to understanding chromosome inheritance since there is no better place to consider mechanisms of inheritance than in the germline. A particular strength of our collaborative team is that we can use our highly resolved structural knowledge of the centromere to help us design advanced genetic and imaging experiments in the mouse germline.
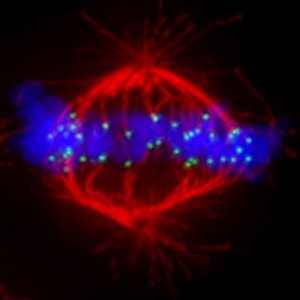
 Mechanisms to avoid mitotic errors in genome transmission
Mechanisms to avoid mitotic errors in genome transmission
While the centromere is the locus that directs chromosome transmission at cell division, the kinetochore is the massive proteinaceous structure that mediates attachment to the microtubule-based spindle. Prior to segregation, sister kinetochores are bound by microtubules emanating from opposite spindle poles. Misconnections must be selectively destabilized by the action of the Aurora B kinase, which phosphorylates kinetochore targets to reduce microtubule binding. Some key areas where we are making important advances include: 1) sub-centromeric mapping of the relevant error corrections complexes on mitotic chromosomes, 2) dissecting the roles of other regulatory molecules upstream and downstream of Aurora B, and 3) atomic-level interrogation of how Aurora B achieves the high level of catalytic activity required for effective error correction.
DNA damage detection and the poly-ADP ribose polymerase: PARP-1
Outside of mitosis, a major threat to the integrity of our genome is the persistent breakage that occurs. The breaks must be rapidly detected, and the ‘first-responder’ to a DNA break is the poly-ADP ribose polymerase, PARP-1. It signals the DNA break by catalyzing the synthesis of ADP ribose polymers from substrate, NAD+. Holding PARP-1 at bay in the absence of DNA damage is critical, and persistent PARP-1 hyperactivity leads to cell death in diverse human diseases. In healthy cells, PARP-1 undergoes a ~1000-fold catalytic activation when it directly recognizes and binds to broken DNA. We are employing HXMS to uncover the physical basis for this activation and to form the basis for future efforts to modulate PARP-1 activity. We are also interested to learn how PARP-1 activity is modulated outside of the DNA damage response where it is known to be involved in the regulation of gene expression.


